1kb Opti-DNA Marker
| Cat# | Unit |
|---|---|
| G016 | 500 μl/100 loads |
Documents
Description
Description
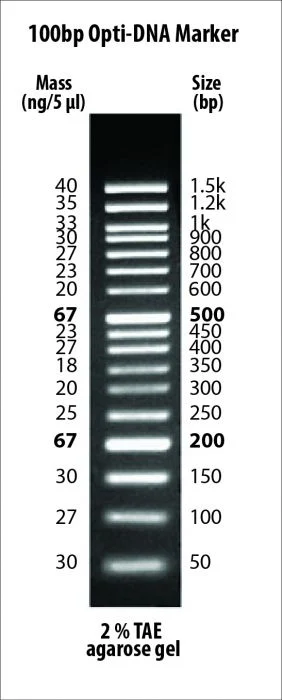
| Description | PCR products and double-stranded DNA were digested with appropriate restriction enzymes to completion to yield 19 bands, ranging from 100 pb – 10 kb, for molecular weight standards in agarose gel electrophoresis. The 500, 1,500, and 3,000 base pair bands have increased intensity to serve as quick reference points. Approximated mass of each DNA band is provided (for a loading size of 5 μl of the DNA ladder) for approximating the mass of DNA in comparably intense DNA samples of similar size. |
|---|---|
| SKU | G106 |
| Applications | For approximating DNA size and amount on agarose gels. |
| Bands | 19 |
| DNA Range | 100bp-10kb |
| Note | Concentration of this marker is 172 μg / ml |
| Suggested Load | Mix gently by pipetting up and down a few time, then directly load 5 μl into well of gel. |
| Shipping Conditions | Shipped on blue ice packs. |
| Storage Condition | Store @ 4°C for up to 3 months or -20°C for up to 24 months. |
| Unit Quantity | 500 μl/100 loads |
Is it ready for use?
Yes, it is ready to use. Just load suggested volume (5ul) to the well.
What is the smallest size agarose gel can separate?
That is around 300bp for 1% agarose and 100bp for 2.5% agarose. With anything smaller than the dye, there may be separation but it will be difficult to tell when the fragment will run off the gel.
Are the Opti-DNA markers compatible with radiolabelling?
Our lab has not tested the Opti-DNA markers for this particular application, but we believe it shouldn’t be a problem. The Opti-DNA markers should be compatible for radio-labelling.
- Gnanandarajah, JS et al. “Identification by mass spectrometry and immune response analysis of guinea pig cytomegalovirus (GPCMV) pentameric complex proteins GP129, 131 and 133” Viruses 6 (2):727-751 (2014). DOI: 10.3390/v6020727. PubMed: 24531333. Application: PCR Products size reference.
- Ly, T. T. M., & Yen, D. T. “Differentiation of two Pangasius species, Pangasius krempfi and Pangasius mekongensis using inter- simple sequence repeat markers” International Journal of Fisheries and Aquatic Studies 7(4):116–120 (2019).