Mycoplasma PCR Detection Kit
| Cat# | Unit |
|---|---|
| G238 | 100 rxn |
Documents
Description
Description
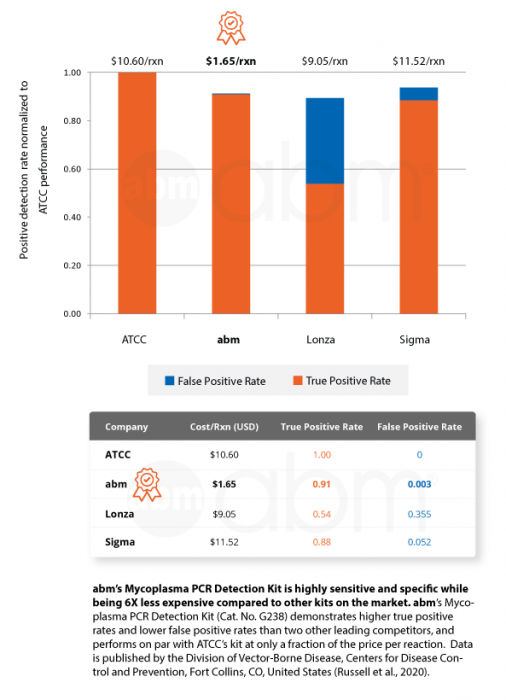
Enjoy superior performance at lower costs!
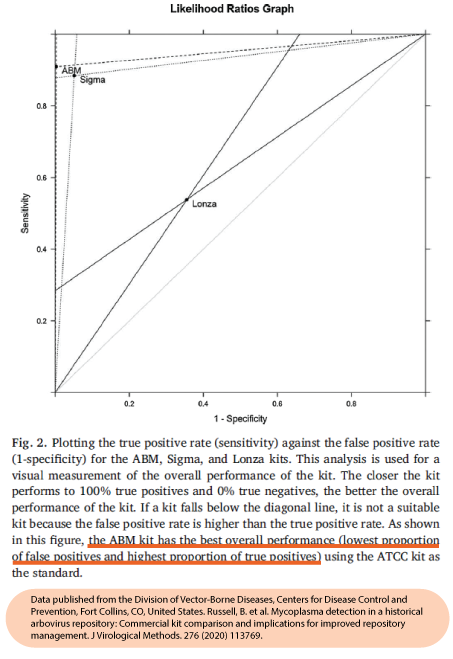
abm‘s Mycoplasma PCR Detection Kit (Cat. No. G238) offers highly specific and sensitive detection of 200+ strains of Mycoplasmas in less than 2 hours. This bestselling kit is designed to minimize false positives, ensuring quick and reliable routine screening of cell cultures. A recent official publication by a national public health agency found that our kit out-performs Sigma and Lonza’s kits while being 6X less expensive than ATCC’s kit (Russell et al., 2020).
Mycoplasmas are highly undesirable, easily acquired, and notoriously difficult to detect, altering infected cells at the molecular level and leading to visible changes in cell morphology and growth characteristics. Timely detection of Mycoplasmas in cell cultures is recommended in order to deter wide-spread contamination and save on the costly efforts of elimination. Our proprietary MasterMix formulation contains a gel loading dye, making it convenient for gel electrophoresis.
Kit Features:
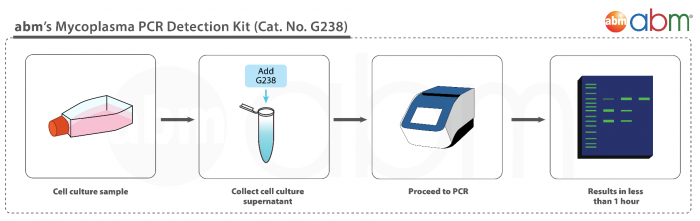
- No DNA isolation needed: Directly add cell culture supernatants to PCR reaction
- Rapid and Comprehensive: Detect 200+ Mycoplasma species in less than 2 hours
- Highly Sensitive and Specific: Demonstrated the highest true positive and lowest false positive rates compared to competing kits
- Easy-to-use: Includes ready-to-use primer mix and positive control
- 6X less expensive per sample compared to competing kits (Russel et al., 2020)
- Out-performs Sigma and Lonza’s kits (Russel et al., 2020)
- Combine with our bestselling Mycoplasma Elimination Cocktail (Cat. No. G398) to eliminate 70+ species of Mycoplasma
| PRODUCT COMPONENTS | QUANTITY |
|---|---|
| BlasTaq™ 2X PCR MasterMix |
1.25 ml
|
| Primer Mix |
100 μl
|
| Positive Control |
250 μl
|
| Nuclease-Free H2O |
1.0 ml
|
Why test your cells for Mycoplasma?
Due to its small size, Mycoplasma can be present in your growth media without being visibly detected. This can cause significant errors in your experiments due to its effects on:
- proliferation
- metabolism
- gene synthesis & processing
- adhesion properties
- and more
Many publishers now request Mycoplasma testing to be reported along with manuscript submissions to ensure accuracy in data reporting.
| SKU | G238 |
|---|---|
| Storage Condition |
Store at -20°C. This product is stable for 2 years from the date of shipping if stored and handled properly. |
| Unit quantity | 100 rxn |
Do you provide or sell a positive control so that we can be sure the assay worked?
Yes, we do have a positive control included in the kit.
What size plate, dish or flask should I use for the culturing of my cells?
We recommend culturing your cells in a 6-well plate or 10cm dish.
What is the basis of the positive control in the mycoplasma kit?
It is DNA fragments of Mycoplasma gDNA diluted in medium.
Can tissue culture supernatant be frozen and thawed for the test?
Yes
I kept the cells in the presence of gentamycin, Should I use antibiotics-free media to detect mycoplasma in the media?
It is not necessary to remove routine antibiotics (e.g. penicillin, streptomycin, and gentamicin) from the media prior to PCR detection, as they will not interfere with the assay.
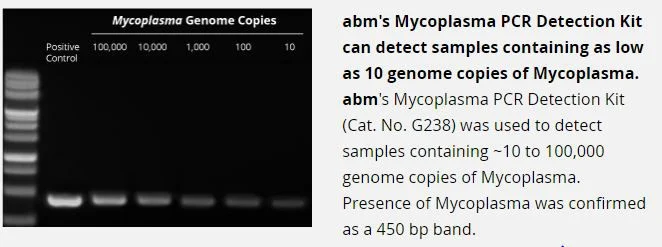
What is the detection limit of your kit?
Our kit is able to detect as low as 10 copies of Mycoplasma /sample
Our cells are grown in suspension and would not achieve 80% confluence, is there a target cell density (cells/ml) we should achieve to ensure we will detect the mycoplasma?
Mycoplasma testing is more dependent on how recently the cells have been subcultured. Seeding density does not play a major role when using this kit. We recommend cells be around at least 3-5 days old in culture without changing any fresh media so as to detect secreted mycoplasma in culture supernatant (i.e. the cell culture medium should not be refreshed/changed for 48-72 hours prior to collection).
Can I use cryogenically frozen supernatant?
Using samples stored at -80C should be fine for this kit. If you thawing out the cells and performing mycoplasma testing right away, usually when thawing the cells the supernatant from the vial is diluted when adding to the cell culture vessel. As long as the DMSO is diluted and the DMSO content is less than 0.5% final in the PCR reaction it should not cause a problem. Too much DMSO would alter the Tm of the PCR buffer and thus affect amplification.
Since I have such a large number of cell lines to test, can I store the cell culture supernatant prior to use in the PCR, or does it have to be freshly collected?
In general, the more fresh the sample is when running the PCR, the better; in keeping with this, it is normally advised to wait until all cell lines are ready to go prior to collecting the supernatant for each. However, if necessary, you can certainly store collected supernatant at +4C storage for short-term (1-2 days) storage; note that we highly recommend avoiding repeated freeze thaw cycles.
How many days after thawing my cells can I perform this mycoplasma test?
We recommend the cells should remain in culture for at least 48-72 hours prior to screening for the presence of mycoplasmas, and that the media sample collection only be done once the cells have reached at least 80% confluence.
There is a ~200-250bp band in one of my samples, does this mean it is mycoplasma positive?
In rare cases, there may be a weak non-specific binding to a region of human gDNA due to the degenerate nature of the primers that can amplify a weak 250bp band in human cell samples. Your sample can be considered as negative for mycoplasma, as mycoplasma amplicons will be 370-550bp in size. You can try to increase the annealing temperature of the PCR program to 60°C to reduce this non-specific amplification.
I did not see amplification of the positive control. What can I try?
This could occur in cases where the PCR machine does not ramp temperature fast enough. We suggest trying touchdown PCR in these situations.
- Koh, JH et al. “Effect of water-soluble fraction of cherry tomatoes on the adhesion of probiotics and Salmonella to intestinal epithelial cells” J Sci Food Agric : (2013). PubMed: 23749725. Application: Cell Culture.
- Jagadish, N et al. “A-kinase anchor protein 4 (AKAP4) a promising therapeutic target of colorectal cancer” J Exp Clin Cancer Res 34(1):142 (2015). DOI: 10.1186/s13046-015-0258-y. PubMed: 26590805. Application: Cell Culture.
- Saha, A et al. “Effect of Metformin, Rapamycin, and Their Combination on Growth and Progression of Prostate Tumors in HiMyc Mice.” Cancer Prev Res (Phila) 8(7):597-606 (2015). DOI: 10.1158/1940-6207.CAPR-15-0014. PubMed: 25908508. Application: Cell Culture.
- Jagadish, N et al. “Heat shock protein 70–2 (HSP70-2) is a novel therapeutic target for colorectal cancer and is associated with tumor growth” BMC Cancer 1:561 (2016). DOI: 10.1186/s12885-016-2592-7. Application: Mycoplasma detection.
- Blouin, M. J., Parisotto, M., Papavasiliou, V., Lavoie, C., Larsson, O., Ohh, M., … & Ferbeyre, G. “Translational and HIF-1a-Dependent Metabolic Reprogramming Underpin Metabolic Plasticity and Responses to Kinase Inhibitors and Biguanides” Cell Metabolism 28:1-16 (2018).
- Bufalieri, F., Infante, P., Bernardi, F., Caimano, M., Romania, P., Moretti, M., … Di Marcotullio, L. “ERAP1 promotes Hedgehog-dependent tumorigenesis by controlling USP47-mediated degradation of βTrCP” Nature Communications 10(1): (2019). DOI: 10.1038/s41467-019-11093-0.
- Carter, B. Z., Mak, P. Y., Wang, X., Tao, W., Ruvolo, V., Mak, D., … Andreeff, M. “An ARC-Regulated IL1β/Cox-2/PGE2/β-Catenin/ARC Circuit Controls Leukemia–Microenvironment Interactions and Confers Drug Resistance in AML” Cancer Research 79(6):1165–1177 (2019). DOI: 10.1158/0008-5472.can-18-0921.
- Chakravorty, S., Yan, B., Wang, C., Wang, L., Quaid, J. T., Lin, C. F., … & Chopra, G. “Integrated Pan-Cancer Map of EBV-Associated Neoplasms Reveals Functional Host–Virus Interactions” Cancer research. : (2019).
- Guarino, A.M., Mauro, G.D., Ruggiero, G. “doi:10” 1038/s41598-019-45468-6 : (2019). DOI: 10.1038/s41598-019-45468-6.
- Hsu, H. P., Wang, C. Y., Hsieh, P. Y., Fang, J. H., & Chen, Y. L. “Knockdown of serine/threonine-protein kinase 24 promotes tumorigenesis and myeloid-derived suppressor cell expansion in an orthotopic immunocompetent gastric cancer animal model” Journal of Cancer 11(1):213-228 (2020).
- Hsueh, Y. S., Chang, H. H., Shan, Y. S., Sun, H. S., Fletcher, J. A., Li, C. F., & Chen, L. T. “Nuclear KIT induces a NFKBIB-RELA-KIT autoregulatory loop in imatinib-resistant gastrointestinal stromal tumors” Oncogene 38(38):6550-6565 (2019).
- Ide, H., Inoue, S., Mizushima, T., Kashiwagi, E., Zheng, Y., & Miyamoto, H. “Role of glucocorticoid signaling in urothelial tumorigenesis: Inhibition by prednisone presumably through inducing glucocorticoid receptor transrepression” Molecular carcinogenesis. : (2019).
- Inoue, S., Ide, H., Mizushima, T., Jiang, G., Netto, G. J., Gotoh, M., & Miyamoto, H. “Nuclear Factor-κB Promotes Urothelial Tumorigenesis and Cancer Progression via Cooperation with Androgen Receptor Signaling” Molecular Cancer Therapeutics 17(6):1303–1314 (2018). DOI: 10.1158/1535-7163.mct-17-0786.
- Inoue, S., Mizushima, T., Ide, H., Jiang, G., Goto, T., Nagata, Y., … Miyamoto, H. “ATF2 promotes urothelial cancer outgrowth via cooperation with androgen receptor signaling” Endocrine Connections 1397–1408: (2018). DOI: 10.1530/ec-18-0364.
- Jones, P. S., Yekula, A., Lansbury, E., Small, J. L., Ayinon, C., Mordecai, S., … & Ghiran, I. “Characterization of plasma-derived protoporphyrin-IX-positive extracellular vesicles following 5-ALA use in patients with malignant glioma” EBioMedicine. : (2019).
- Kollara, A., Shathasivam, P., Park, S., Ringuette, M. J., & Brown, T. J. “Increased Androgen Receptor Levels and Signaling in Ovarian Cancer Cells by VEPH1 Associated with Suppression of SMAD3 and AKT Activation” The Journal of Steroid Biochemistry and Molecular Biology 105498: (2019).
- Lee, R. S., Zhang, L., Berger, A., Lawrence, M. G., Song, J., Niranjan, B., … & Risbridger, G. P. “Characterization of the ERG-regulated Kinome in Prostate Cancer Identifies TNIK as a Potential Therapeutic Target” Neoplasia 21(4):389-400 (2019).
- Linley, A. J., Griffin, R., Cicconi, S., D’Avola, A., MacEwan, D. J., Pettitt, A. R., … & Slupsky, J. “Kinobead profiling reveals reprogramming of B-cell receptor signaling in response to therapy within primary CLL cells” bioRxiv 841312: (2019).
- Lospinoso Severini, L., Quaglio, D., Basili, I., Ghirga, F., Bufalieri, F., Caimano, M., … & Maroder, M. “A Smo/Gli Multitarget Hedgehog Pathway Inhibitor Impairs Tumor Growth” Cancers 11(10):1518 (2019).
- Mehrpour, M., Rodriguez, R., Hamai, A., & Trang, M. “U” S. Patent Application No. 16/082 970: (2019).
- Nii, T., Prabhu, V.V, Ruvolo, V., Madhukar, N., Zhao, R., Mu, H., Heese, L., Nishida, Y., Kojima, K., Garnett, M.J., McDermott, U., Benes, C.H., Charter, N., Deacon, S., Elemento, O., Allen, J.E., Oster, W., Stogniew, M., Ishizawa, J., Andreeff, M. “Imipridone ONC212 activates orphan G protein-coupled receptor GPR132 and integrated stress response in acute myeloid leukemia” Leukemia. : (2019). DOI: 10.1038/s41375-019-0491-z.
- Ong, F., Dreesen, O., & Dröge, P. . “The RNA interactome of human telomerase RNA reveals a coding-independent role for 2 a histone mRNA in telomere homeostasis 3” : ().
- Rosenberger, L., Ezquer, M., Lillo-Vera, F., Pedraza, P. L., Ortúzar, M. I., González, P. L., … Alcayaga-Miranda, F. “Stem cell exosomes inhibit angiogenesis and tumor growth of oral squamous cell carcinoma” Scientific Reports 9(1): (2019). DOI: 10.1038/s41598-018-36855-6.
- Ross, J., Bressler, K., & Thakor, N. “Eukaryotic Initiation Factor 5B (eIF5B) Cooperates with eIF1A and eIF5 to Facilitate uORF2-Mediated Repression of ATF4 Translation” International Journal of Molecular Sciences 19(12):4032 (2018). DOI: 10.3390/ijms19124032.
- Salvati, A., Gigantino, V., Nassa, G., Giurato, G., Alexandrova, E., Rizzo, F., … Weisz, A. “The Histone Methyltransferase DOT1L Is a Functional Component of Estrogen Receptor Alpha Signaling in Ovarian Cancer Cells” Cancers 11(11):1720 (2019). DOI: 10.3390/cancers11111720.
- Severini, L. L., Quaglio, D., Basili, I., Ghirga, F., Bufalieri, F., Caimano, M., … & Maroder, M. “A Smo/Gli Multitarget Hedgehog Pathway Inhibitor Impairs Tumor Growth” Cancers 11(10): (2019).
- Shishodia, G., Koul, S., & Koul, H. K. “Protocadherin 7 is overexpressed in castration resistant prostate cancer and promotes aberrant MEK and AKT signaling” The Prostate 79(15):1739–1751 (2019). DOI: 10.1002/pros.23898.
- Singh, S., Kumar, M., Kumar, S., Sen, S., Upadhyay, P., Bhattacharjee, S., … & Kundu, T. K. “The cancer-associated, gain-of-function TP53 variant P152Lp53 activates multiple signaling pathways implicated in tumorigenesis” Journal of Biological Chemistry 294(38):14081-14095 (2019).
- Spiombi, E., Angrisani, A., Fonte, S., De Feudis, G., Fabretti, F., Cucchi, D., … & Di Magno, L. “KCTD15 inhibits the Hedgehog pathway in Medulloblastoma cells by increasing protein levels of the oncosuppressor KCASH2” Oncogenesis 8(11):1-11 (2019).
- Wang, D., Luo, H., Huo, Z., Chen, M., Han, Z., Hung, M., … & Xiao, H. “Irradiation-induced dynamic changes of gene signatures reveal gain of metastatic ability in nasopharyngeal carcinoma” American Journal of Cancer Research 9(3):479 (2019).
- Wen, M. H., Xie, X., Tub, J., Lee, D. F., & Chen, T. Y. “Generation of a genetically modified human embryonic stem cells expressing fluorescence tagged ATOX1” Stem Cell Research 101631: (2019).
- Wilson, M. R., Reske, J. J., Holladay, J., Wilber, G. E., Rhodes, M., Koeman, J., … & Patterson, A. L. “ARID1A and PI3-kinase pathway mutations in the endometrium drive epithelial transdifferentiation and collective invasion” Nature communications 10(1):1-18 (2019).
- Yekula, A., Minciacchi, V. R., Morello, M., Shao, H., Park, Y., Zhang, X., … & Carter, B. “Large and small extracellular vesicles released by glioma cells in vitro and in vivo” Journal of Extracellular Vesicles 9(1):1689784 (2019).
- Russell, B. J., Horiuchi, K., Velez, J. O., Goodman, C. H., & Johnson, B. W. (2020). Mycoplasma detection in a historical arbovirus repository: Commercial kit comparison and implications for improved repository management. Journal of virological methods, 276, 113769. https://doi.org/10.1016/j.jviromet.2019.113769