Authorized for Clinical Use Under Emergency Use Authorization
The Coronavirus pandemic led to a rapid response from the viral research and therapeutics community around the world. In partnership with Biotia, Inc., Twist has received Emergency Use Authorization (EUA) from the U.S. Food and Drug Administration (FDA) for the SARS-CoV-2 Next-Generation Sequencing (NGS) Assay. The SARS-CoV-2 NGS Assay is an in vitro diagnostic test, a highly sensitive nucleic acid hybridization capture-based assay, intended for the detection of SARS-CoV-2 RNA. Combined with Biotia’s comprehensive data analysis software and reporting capabilities (COVID-DX (v1.0)), the test reduces the likelihood of a false-negative result. In contrast, a majority of SARS-CoV-2 tests based on polymerase chain reaction (PCR) only identify limited genetic markers of the virus. The underlying hybrid-capture technology has a high variant tolerance to accurately identify and characterize novel or uncharacterized variants, maximizing the number of variant sequences identified, where other methods may miss mutations in certain regions.
The SARS-CoV-2 NGS Assay enables complete viral genome sequencing from a variety of sources (such as nasopharyngeal swabs) with as little as 800 viral copies per milliliter. The assay enables batch analysis of up to 96 samples (92 patient samples concurrently with four controls) in a single test to indicate the presence or absence of the virus in each sample.
In addition to receiving reliable detection results, researchers have the option of obtaining a research-use only (RUO) report at no additional cost. The RUO report includes genetic variants detected, clade-associated variant results, and phylogenetic analysis, all of which allow surveillance of viral mutations, genetic variability, and evolution.
KEY BENEFITS

- Limit of Detection (LoD) determined to be 800 viral copies per milliliter
- Identify variants to reduce the likelihood of false-negative results

- Exceeds 95% FDA concordance guidelines

- Provides variant typing and phylogenetic analysis to enable epidemiological surveillance
*This product has not been FDA cleared or approved, but has been authorized for emergency use by FDA under an EUA for use by authorized laboratories; This product has been authorized only for the detection of nucleic acid from SARS-CoV-2, not for any other viruses or pathogens; and, the emergency use of this product is only authorized for the duration of the declaration that circumstances exist justifying the authorization of emergency use of in vitro diagnostics for detection and/or diagnosis of COVID-19 under Section 564(b)(1) of the Federal Food, Drug and Cosmetic Act, 21 U.S.C. § 360bbb-3(b)(1), unless the declaration is terminated or authorization is revoked sooner.
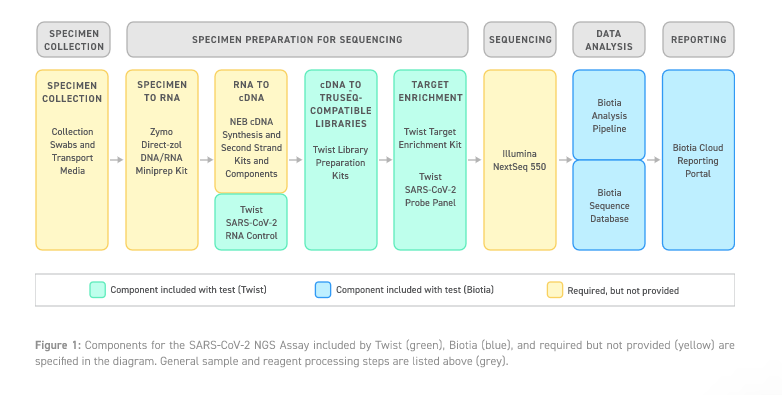
Workflow Overview
The dsDNA biotin-bound hybrid capture panel used in this assay is designed to enrich for all evolved SARS-CoV-2 virus sequences in upper respiratory specimens from patients suspected of COVID-19.
This capture-based assay utilizes cDNA synthesis from template viral and human RNA sequence in a patient’s specimen, producing amplified libraries from the cDNA using a PCR thermocycler. The SARS-CoV-2 virus sequence is then targeted during a hybridization reaction with a biotin-attached probe specific to the SARS-CoV-2 virus. Targeted regions are extracted from the non-targeted regions using biotin-streptavidin chemistry and stringency washes. Remaining molecules are amplified using a PCR thermocycler and sequenced by an Illumina-based NGS platform (NextSeq 500, NextSeq 550, and NextSeq 550Dx system).
Data Analysis and Reporting
The Biotia COVID-DX (v1.0) software analyzes sequencing results to provide a clinically oriented report that includes results on the presence or absence of the SARS-CoV-2 virus in each sample. The software is for diagnostic use under the FDA Emergency Use Authorization. Sample report shown in Figure 2.
Presence/Absence
Detection of SARS-CoV-2 is based on the percentage of the viral genome that is recovered from a patient’s sample. The higher the percentage of viral genome that is recovered, the more likely it is that the assay will determine that the virus is present.
Results interpretation
POSITIVE REPORT: SARS-CoV-2 viral genome was detected.
NEGATIVE REPORT: SARS-CoV-2 viral genome was NOT detected.
INVALID REPORT: controls did not pass filter. Sample should be tested again. If a second failure occurs, it is reported to the sender as “invalid result” and recollection is recommended if patient testing is still clinically indicated.
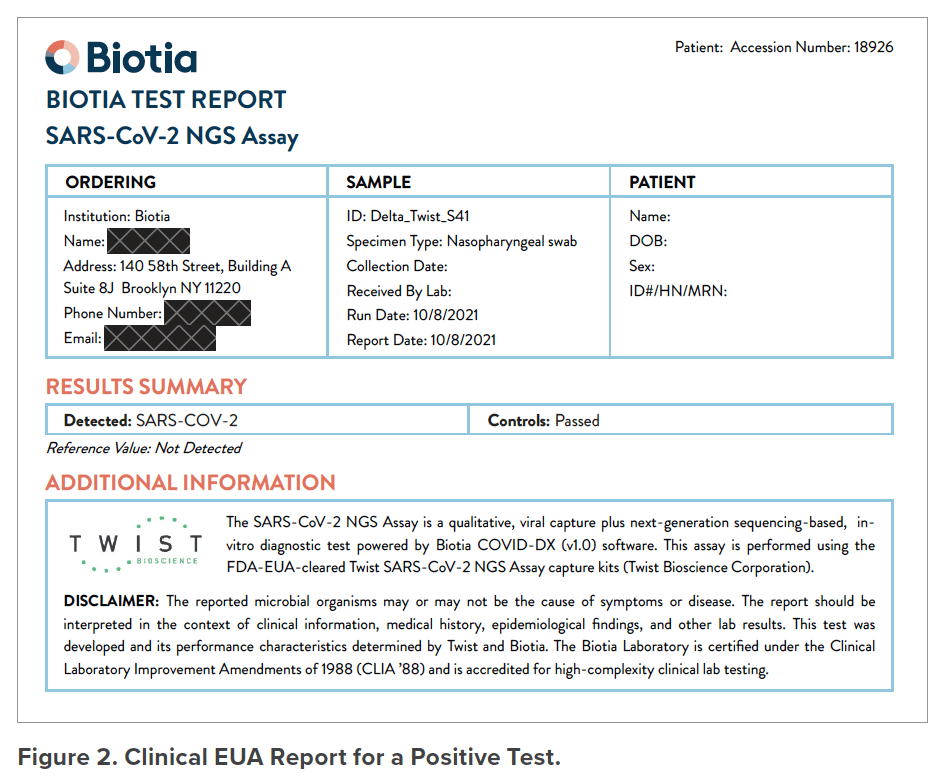
Clinical EUA Report for a Positive Test
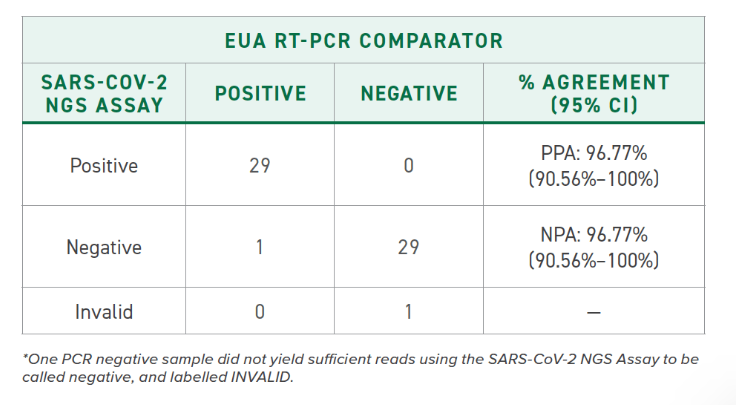
Clinical performance of the SARS-CoV-2 NGS Assay was evaluated by comparing results to an RT-PCR assay authorized by the FDA for use under Emergency Use Authorization (EUA RT-PCR). 60 clinical nasopharyngeal (NP) swab specimens were tested, including 30 SARS-CoV-2-positive and 30 SARS-CoV-2-negative specimens. The positive and negative percent agreement was calculated in relation to the EUA RT-PCR comparator method and is indicated in the following to the right.
Performance Characteristics
Limit of Detection (LoD)
An analytical sensitivity study established the lowest concentration of SARS-CoV-2 (genome copies(cp)/μl) that can be detected by the SARS-CoV-2 NGS Assay at least 95% of the time. The LoD was determined to be 800 copies/ml and was replicated 30 times with 29/30 (96.67%) samples testing positive.
Clinical Evaluation
Clinical performance of the SARS-CoV-2 NGS Assay was evaluated by comparing results to an RT-PCR assay authorized by the FDA for use under Emergency Use Authorization (EUA RT-PCR). 120 clinical nasopharyngeal (NP) swab specimens were tested, including 60 SARS-CoV-2-positive and 60 SARS-CoV-2-negative specimens. The positive and negative percent agreement was calculated in relation to the EUA RT-PCR comparator method and is indicated in the following to the right.
PRODUCT SHEET
PROTOCOL




