The Coronavirus pandemic led to a rapid response from the viral research and therapeutics community around the world. These research teams need robust and reliable genomic tools to classify and characterize viral samples. Twist offers Research-Use Only (RUO) NGS target enrichment panels for the detection and characterization of SARS-CoV-2 virus. The panel targets about 30 kb viral genome with approximately 1,000 probes, designed against the SARS-CoV-2 genome (GenBank: MN908947.3).
KEY BENEFITS

- Detection of as little as 10 copies of viral material with a fold enrichment of >100,000

- Coverage of > 99.9% of the genome at 1X or greater post enrichment
- Ability to identify viral mutations

- Assess viral evolution over time
- Track strain origin and transmission patterns
NGS for Virus Detection
Next-generation sequencing (NGS) offers high-throughput, specific identification of infections in a variety of sample types including blood, nasal swab, feces, etc. In the case of viral infections, however, direct sequencing can be a challenge due to the extremely low concentration of the viruses of interest and the presence of high background from the host. Target capture using DNA-based hybridization probes to isolate specific sequences out of a mixed genomic sample can increase the sensitivity and specificity of NGS-based efforts.
High Sensitivity Through Efficient Design
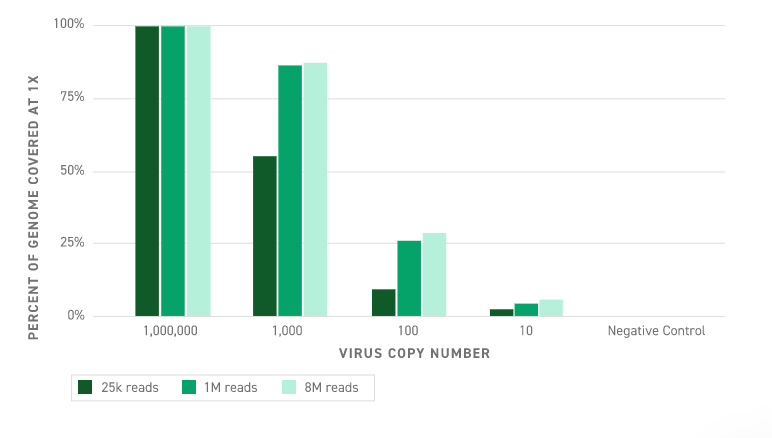
Twist’s highly sensitive and accurate target capture enables efficient viral RNA detection. We demonstrate target enrichment’s ability to capture and detect the viral genome at varying numbers of viral copies spiked into human reference RNA. At high viral load (approximately 1 million copies), the complete genome is recovered even with only 25,000 reads*. While at lower copy numbers, coverage of large portions of the viral template is maintained. No reads from the negative control map to the viral template even when using 8 million mapped reads, demonstrating our detection at 10 copies represents true capture and not background contamination. Find out more in the App Note.
*When using COVID-Dx, 1 million reads are necessary for analytics.
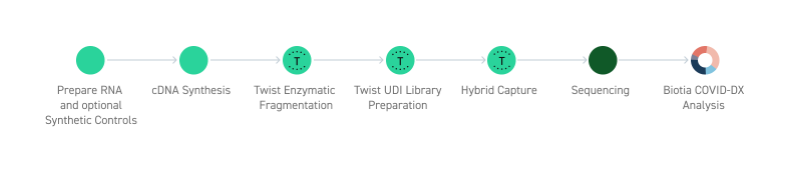
Easy Workflow from Purified RNA to Sequencing
The Twist SARS-CoV-2 Research Panel is provided with a simple-to-follow protocol from purified RNA samples to sequencing with easily available components.
Biotia Analysis
Twist SARS-CoV-2 Research Panel has been combined with additional reagents and analysis to create an end-to-end solution from sample collection through tertiary analytics, providing the SARS-CoV-2 NGS Assay.
The Biotia COVID-DX (v1.0) software provides a research-oriented report that includes the full sequence of the SARS-CoV-2 virus, enabling improved understanding of mutations, genetic variations, and the evolution of the virus as it is transmitted. FASTQ files (sequencing output) can be generated in laboratories and submitted to Biotia COVID-DX (v1.0), a cloud-based software, to generate research-use only reports. The purchase of each SARS-CoV-2 NGS Assay kit includes credits for COVID-DX (v1.0) bioinformatic analysis of each sample.
To receive Biotia analysis credits, simply register through the Biotia User Portal via www.biotia.io and enter the unique order number emailed to you once you place your order. These credits provide access to run samples through the report generation component of the software.
Genetic variants identified
Genetic variants are determined by aligning the test strain to the SARS-CoV-2 reference genome (NC_045512.2) with information on gene location/site, alteration and literature notes if any.
| Detected SARS-CoV-2 | Gene | Site (bp) | Alteration | Reference value |
| Yes | GORF1ab | 241 | T | Not Detected |
| Yes | ORF1ab | 478 | T | Not Detected |
| Yes | ORF1ab | 3,037 | T | Not Detected |
| Yes | ORF1ab | 14408* | T | Not Detected |
| Yes | ORF1ab | 18877 | T | Not Detected |
| Yes | S | 23403 | G | Not Detected |
| Yes | 3a | 25563 | T | Not Detected |
| Yes | 3’UTR | 29759 | C | Not Detected |
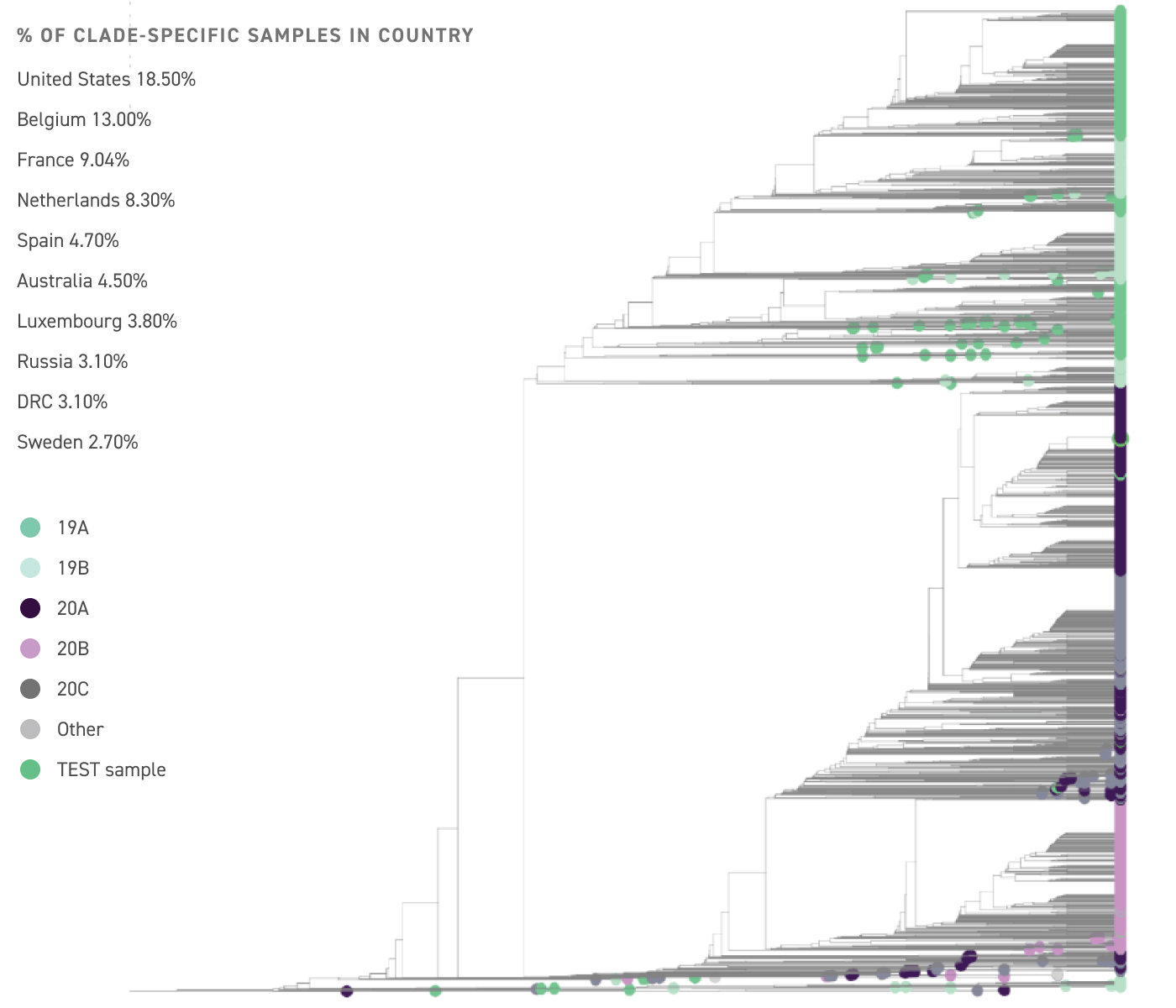
Sequence results mapped across the genome
The percent of the SARS-CoV-2 genome recovered from your test sample, and genetic variants identified compared to the reference genome are indicated. The proportion of known genetic variants of SARS-CoV-2 strains from across the world are shown (bottom).

Phylogenetic analysis and surveillance monitoring.
The test sample is most closely related to Clade 20A
This clade is found with the highest frequency a mong sequenced samples in the United States, Belgium, and France
PRODUCT SKUS – LIBRARY PREP + REAGENTS + RESEARCH PANEL + BIOTIA
104796: Twist SARS-CoV-2 NGS Assay – RUO, 16 Samples
104797: Twist SARS-CoV-2 NGS Assay – RUO, 96 Samples
104799: Twist SARS-CoV-2 NGS Assay – RUO, 768 Samples
PRODUCT SKUS – RESEARCH PANEL + BIOTIA
103564: Twist SARS-CoV-2 Research Panel + Biotia COVID DX (v1.0) – RUO, 2 Reactions, Kit
103566: Twist SARS-CoV-2 Research Panel + Biotia COVID DX (v1.0) – RUO, 12 Reactions, Kit
103567: Twist SARS-CoV-2 Research Panel + Biotia COVID DX (v1.0) – RUO, 96 Reactions, Kit
PRODUCT SKUS – RESEARCH PANEL
102016: Twist SARS-CoV-2 Research Panel, 2 Reactions, Kit
102017: Twist SARS-CoV-2 Research Panel, 12 Reactions, Kit
102018: Twist SARS-CoV-2 Research Panel, 96 Reactions, Kit
The Twist Synthetic SARS-CoV-2 RNA Controls are for research use only, and subject to additional use restrictions as set forth in Twist’s Supply Terms and Conditions.




